Research
Overview of Bhattacharyya lab research projects
The Bhattacharyya lab focuses on characterizing the response of pathogens (bacteria and fungi) to antimicrobials, and of patients to systemic infections. While we use many approaches to address these broad topics, our "bread and butter" is a combination of standard microbiological methods and transcriptional profiling. As an early manifestation of cellular responses to stimuli, gene expression profiles, particularly in response to key perturbations like antibiotic exposure or infection, reflect physiological adaptations by both microbes and patients that we can use to infer mechanisms of disease pathogenesis, and also co-opt for diagnostic purposes. Facilitating this strategy, technological advances in nucleic acid sequencing enable systematic, unbiased, high-throughput transcriptomic profiling of pathogens, and of human cells at single-cell resolution, informing our understanding of the crucial interplay between pathogens, antibiotics, and the human immune system (Fig 1, below). To download an overview of projects in the lab as of Sep 2023, check out our lab poster from Harvard BBS graduate student orientation.Figure 1. Transcriptomic profiling of bacteria upon antibiotic exposure (above), and of human immune cells at single-cell resolution in systemic infections (below), can inform our understanding of these important states and be co-opted for clinical diagnostics.
1. A novel approach to clinical antimicrobial susceptibility testing (AST):
Motivation: The current gold standard of growth-based AST requires several days to return answers. Thus, initial antibiotics must be chosen empirically, leading to overuse of broad-spectrum antibiotics in cases where targeted agents would be more effective. Diagnostics that inform patient care in real time would dramatically change this dynamic, eliminating the tension between individual and society, improving the efficiency of antibiotic use, and enabling the development and targeted administration of new narrow-spectrum antibiotics.
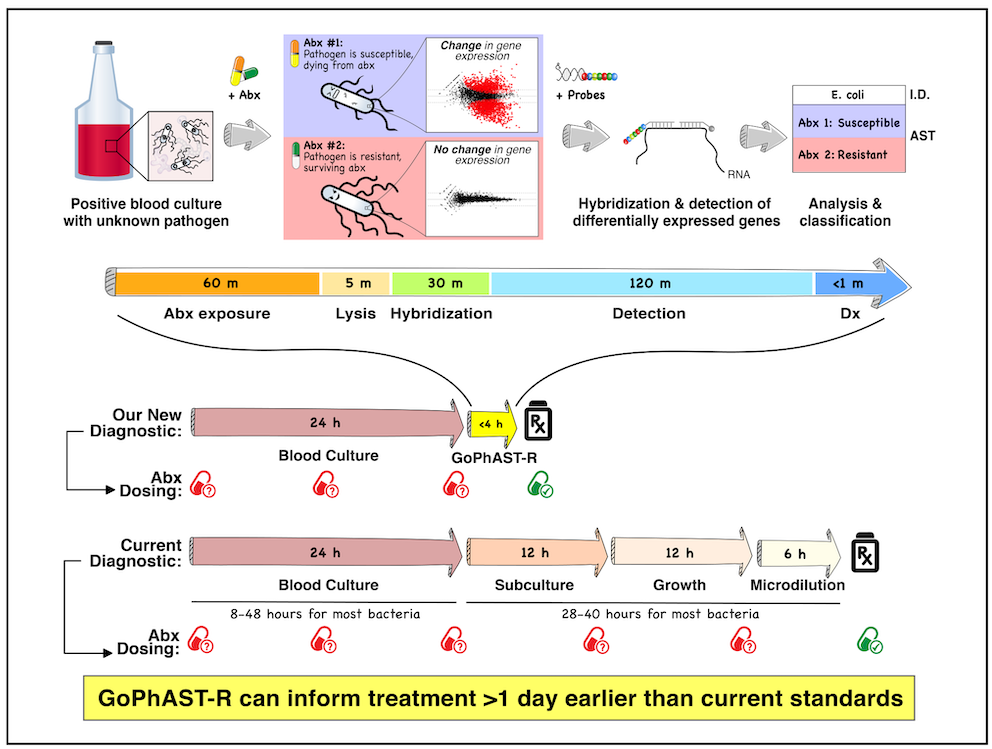
Key findings: Recent diagnostic efforts aimed at genetic AMR markers are limited by incomplete knowledge of diverse microbial resistance mechanisms. We developed an approach to genotypic and phenotypic AST through RNA detection (GoPhAST-R), built on the fact that susceptible strains exhibit major transcriptional changes upon antibiotic exposure, while resistant strains do not, agnostic to resistance mechanism: essentially, dying (or growth-arrested) cells look transcriptionally distinct from those not fated to die, often within minutes of antimicrobial exposure. Coupling these phenotypic transcriptional signatures with genetic resistance determinants on an easy-to-use, commercially available multiplexed RNA detection platform (NanoString) enhances diagnostic accuracy, provides molecular epidemiology, and flags discrepant results for further study, returning results within hours of a positive blood culture, >1 day faster than current assays (Fig 2, below).
Ongoing projects: My lab is extending this new diagnostic paradigm to additional antibiotics, and to more bacteria as well as fungi, ultimately aiming to build this approach into a pan-microbial diagnostic platform that could revolutionize infectious disease diagnostics. In addition, we are using these transcriptional data upon antimicrobial exposure, coupled with microbiological and biochemical measurements, to explore the molecular mechanisms of bacterial killing by, and adaptation to, key antibiotic classes. We have also shown that on the same RNA detection platform, we can identify both bacteria and fungi with near-single-microbe sensitivity with a carefully-designed probeset targeting variable regions of highly abundant rRNA, an approach we call phylogeny-informed rRNA-based strain identification (Phirst-ID); because it is based only on hybridization and requires no enzymology, Phirst-ID can identify pathogens within hours directly from primary clinical samples.
Project leaders: Melanie Martinsen (adapting GoPhAST-R to other antibiotics), Michelle Matzko & Melanie (adapting GoPhAST-R and Phirst-ID to fungi)
Key collaborators: Virginia Pierce (MGH Clinical Microbiology), Christina Cuomo (Broad Fungal Genomics), Poppy Sephton-Clark (Broad Fungal Genomics)
Figure 2. Melanie Martinsen’s cool schematic explaining how transcriptional profiling after antimicrobial exposure can be exploited to distinguish susceptible from resistant pathogens, accelerating AST.
2. Single-cell transcriptional profiling of circulating immune cells in infected patients to improve diagnostics and mechanistic understanding of immune-mediated pathology in systemic infection:
Motivation: The human immune system responds in a nuanced manner to various types of infectious and inflammatory states. Better understanding the immune response to infection is particularly crucial for life-threatening conditions such as sepsis and COVID-19, in which a dysregulated immune response is thought to contribute to morbidity and mortality. The recent revolution in single-cell RNA-Seq (scRNA-Seq) has transformed our understanding of eukaryotic cell types and states, revealing previously unknown heterogeneity at unprecedented detail. We hypothesize that scRNA-Seq of peripheral blood mononuclear cells (PBMCs) can refine transcriptional predictors of infection, improving diagnostic performance of these transcriptional markers and revealing detailed biology underlying the host immune response to infection and infectious mimics.
Key findings: With collaborators at MGH (Drs. Nir Hacohen, Marcia Goldberg, and Michael Filbin) and the Broad Institute (Dr. Paul Blainey and Miguel Reyes), we used scRNA-Seq of PBMCs to identify a novel transcriptional substate of monocytes (“MS1”, or monocyte substate 1) that is enriched in patients with sepsis (Fig 3, below). Based on our analysis of publicly available scRNAseq data, we also find that MS1 is enriched in severe COVID-19. This substate, readily detectable by flow cytometry (Fig 3d), outperformed classical cell states in classifying severity of bacterial infection or COVID-19, provided cellular context for transcriptional signatures of sepsis, and was found in publicly available transcriptional studies of sepsis and of COVID-19, validating our findings.
Ongoing projects: We are building on these findings to better distinguish sepsis from non-infectious critical illness, which is crucial in guiding initial patient management. Both states exhibit MS1, but our preliminary data indicate differential expression of key genes within MS1 cells in sepsis versus other causes of critical illness. We are also exploring the kinetics of the immune response to sepsis during initial treatment, correlated with clinical outcomes and the appropriateness of empiric antibiotic selection, to see if MS1 or other cellular or transcriptional states might serve as prognostic biomarkers in sepsis and other systemic infection that might predict either primary treatment failure or relapsed infection in order to more rationally guide antibiotic selection and duration. We also hope this line of inquiry will better delineate the immune dysregulation that contributes to sepsis pathophysiology.
Project leader: Pierre Ankomah
Key collaborators: Nir Hacohen (MGH & Broad), Marcia Goldberg (MGH ID), Mike Filbin (MGH EM), Paul Blainey (Broad & MIT), Miguel Reyes (MIT), Abraham Sonny (MGH)
Figure 3. MS1 monocytes are enriched in sepsis. (a) The fraction of MS1 cells (relative to total CD45+ cells) is higher in patients with sepsis (URO, SEP) than in healthy controls or those with urinary tract infections with leukocytosis but no signs of sepsis (Leuk-UTI). Asterisk = false discovery rate (FDR) ≤ 0.003 by Wilcoxon rank-sum test. (b) t-distributed stochastic neighbor embedding (tSNE) plot of monocytes colored by embedded density of cells from sepsis patients (left; dark = enriched in sepsis), or by cell state (right), showing enrichment of MS1 in sepsis. (c) Top marker genes differentially expressed (FDR < 0.05 by Wilcoxon rank-sum test) in MS1 cells relative to other cells. (d) Flow cytometry readily identifies MS1 via cell surface markers (low HLA-DR and high IL1R2 in CD14+ monocytes), demonstrating marked enrichment in patients with urosepsis (URO).
3. Resistance to carbapenems and other beta-lactams and the inoculum effect:
Based on a series of observations from the clinical microbiology lab and some of our transcriptional experiments, we are revisiting a long-described issue in beta-lactam resistance, that of the inoculum effect: essentially, the more cells you treat with beta-lactams, the more antibiotic it takes to kill them. We are exploring both the mechanistic basis for this phenomenon (is it simply due to secreted enzymes building up in solution, a form of cooperativity between cells at high density? or is there more to it?) and its clinical consequences (strains with the capacity for high-level resistance may “fool” standard assays done at lower inocula) using carbapenems as a model beta-lactam, given the clinical importance of this ultra-broad-spectrum antibiotic.
Project leaders: Alex Jaramillo & Kyra Taylor (experimental); Pierre Ankomah (modeling)
Key collaborators: Ashlee Earl (Broad Bacterial Genomics), Virginia Pierce (MGH Clin Micro)
4. Genotypic and functional characterization of “interesting” clinical isolates:
Though we do not have a systematic project built around this, Roby occasionally hears about a case from his colleagues on the wards that involves an unusual clinical phenotype or other properties of a strain or infection (or cluster of infections) that piques his interest. In these cases, if someone in lab is interested, we will (with IRB approval and in collaboration with clinical colleagues) investigate the isolates in question, usually though a mix of whole-genome sequencing and functional phenotypic assays, starting with both molecular and conventional microbiological approaches and following our curiosity. While this is nobody’s main project at first, it can be fun and interesting and may even give rise to a more robust project, such as the Citrobacter isolates that first led to Alex and Kyra’s interest in revisiting inoculum effects.
Project leaders: whoever’s interested at the time Roby hears about a cool case!
Key collaborators: MGH Infectious Diseases Division and Clin Micro lab, usually